Projects

Deep sea coral from a seamount in the Marshall Islands (NA174)
Biodiversity Exploration of Seamounts and Other Deep Sea Habitats with eDNA and New Technologies
Seamounts, or underwater mountains, are hotspots of biodiversity in the deep sea. However, seamount animals and their distributions and ecology are poorly studied because the environment is difficult to access. I am leading research funded by the NOAA Ocean Exploration Cooperative Institute to study deep sea seamount habitats using eDNA, autonomous sampling, ROVs, and landers (the Deep Autonomous Profiler). To date, we have sampled seamounts off of Hawaii (NA155), American Samoa (NA164 and NA65), and the Republic of the Marshall Islands (NA174)
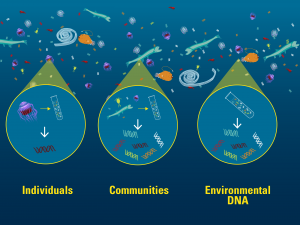
Individual animals, bulk animal collections, and environmental DNA can all be genetically barcoded. Image credit: Govindarajan and Renier, WHOI
Mesopelagic Biodiversity
The vast mesopelagic zone, or twilight zone, is a poorly explored ocean region ranging from 200 to 1000 meters below the ocean surface. I am leading the Biodiversity portion of WHOI's Ocean Twilight Zone project. My work emphasizes the use of molecular approaches such as DNA barcoding and metabarocoding of individuals, plankton tow contents, and eDNA (described below). The animals I am studying include a variety of fish and invertebrates such as jellyfish and crustaceans. I am looking at how animal distributions change over space and time, and particularly the phenomenon of diel vertical migration (DVM). DVM plays an important role in the carbon cycle, because it can facilitate the transfer of carbon from surface waters to the deep sea.

Analyzing traces of genetic material in seawater to detect biodiversity. Photo by Tom Kleindist, WHOI
Using Environmental DNA (eDNA) for biodiversity assessments
Environmental DNA (eDNA) refers to any DNA in the environment, and includes DNA shed by animals through a variety of mechanisms (e.g., spawning, defecation, etc). We can collect eDNA, instead of the animals themselves, to discover what species were present in the area. This approach is powerful, because we can detect animals that traditional sampling methods miss. My laboratory is using eDNA analyses to study the ocean's twilight zone and other marine habitats. For more information, see this primer on eDNA and this Oceanus article.
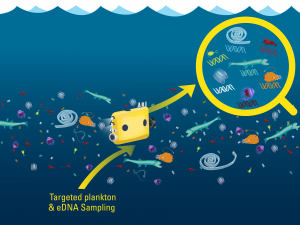
A conceptual image of eDNA sampling on Mesobot. Image credit: Govindarajan and Renier, WHOI
Scaling up and diversifying midwater eDNA sampling strategies using autonomous environmental DNA sampling with in situ filtration
I am co-developing autonomous environmental DNA samplers specifically targeting eDNA from the Ocean's Twilight Zone. I incorporated an eDNA sampler developed in my laboratory on the towed broadband acoustics and imaging instrument, Deep See. You can read more about it here. In 2019 deployed a new, large-volume eDNA multisampler that I co-developed on the robotic vehicle Mesobot in the Gulf of Mexico. The samplers filter water in situ, which is a great advantage over traditional eDNA sampling. We now have a new version of our eDNA samplers (co-developed by Oceanic-Labs and WHOI; Adams, Govindarajan, and Yoerger, in prep.), that we have deployed on both Mesobot and Deep-See. We are expanding to include eDNA samplers on other platforms, including ROVs and a benthic lander. We are also expanding our sampling strategies to include adaptive sampling in response to real-time acoustic backscatter data from the platform itself, the ship, or even another autonomous vehicle.

Images of mesopelagic animals. Reference barcodes for these and other species will enable DNA barcoding analyses, including eDNA analyses. Images by Paul Caiger and Larry Madin.
Reference libraries for DNA barcoding analysis
A major obstacle to DNA barcoding and metabarcoding analysis of samples and eDNA is the lack of reference sequences for taxonomic assignment. As part of the OTZ project, my laboratory is barcoding fish and invertebrate specimens obtained from midwater trawls and MOCNESS sampling. It is critical that the animals used to provide DNA for reference barcodes are accurately identified.