New methods to characterize marine organic matter
Marine organic matter is a complex collection of reduced carbon compounds that contain heteroatoms such as oxygen, nitrogen, phosphorus and sulfur. Given the heterogeneous composition of marine organic matter, it is not surprising that full compositional analysis has been elusive and that analytical challenges for organic matter characterization remain. We are actively improving our ability to understand marine organic matter through the development and application of novel analytical methods.
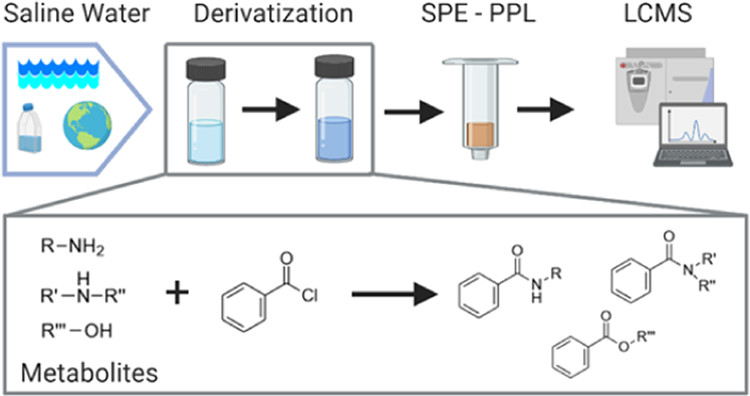
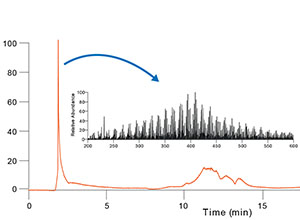
Our method development takes multiple paths. First, we seek to increase our extraction of low molecular weight organic compounds from seawater (see for example Widner et al. 2021). Second, we work to improve the methods used to characterize marine organic matter (see for example Johnson et al. 2017). Finally, ultrahigh resolution mass spectrometers produce large and complex datasets and tools such as AutoTuner facilitate data processing of untargeted metabolomics data.
Our sample analysis is all done within the WHOI FT-MS facility, a facility that is directed by Liz Kujawinski and managed by Melissa Kido Soule.
Recent publications
Widner, B., M. C. Kido Soule, F. Ferrer-González, M. A. Moran and E. B. Kujawinski (2021). Quantification of amine- and alcohol-containing metabolites in saline samples using pre-extraction benzoyl chloride derivatization and ultra-high performance liquid chromatography tandem mass spectrometry (UHPLC MS/MS). Analytical Chemistry. 93: 4809-4817 (link to publication)
McLean, C. and E. B. Kujawinski (2020). AutoTuner: High fidelity, robust, and rapid parameter selection for metabolomics data processing. Analytical Chemistry. 8: 5724-5732 (link to publication)
Johnson W.M., Kido Soule M.C., and Kujawinski E.B. (2017) Interpreting the impact of matrix on extraction efficiency and instrument response in a targeted metabolomics method. Limnology and Oceanography Methods 15: 417-428. link to publication
Longnecker, K. and E. B. Kujawinski (2017). Mining mass spectrometry data: Using new computational tools to find novel organic compounds in complex environmental mixtures. Organic Geochemistry 110:92-99. link to publication