Datasets
Trace Metal Datasets
The CoFeMUG Expedition (Cobalt, Iron, and Microbes from the Upwelling to the Gyre) - R/V Knorr 2007. BCO-DMO data repository. Noble et al, Coastal Plume Limnol. Oceanogr manuscript.
The CORSACS Expedition (Controls on Ross Sea Algal Community Structure) - R/V N.B. Palmer 2005-2006. BCO-DMO data respository. Saito et al., 2010 Biogeosciences Manuscript.
Experimental Microbial Proteomic Datasets
Diel Cycling of Proteins in Crocosphaera watsonii
Saito, MA, EM Bertrand, VV Bulygin, DM Moran, S Dutkiewicz, FM Monteiro, MJ Follows, FW Valois, JB Waterbury. 2011. Iron Conservation by Reduction of Metalloenzyme Inventories in the Marine Diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. doi:10.1073/pnas.1006943108.
Supplemental Proteomics dataset with spectral counting results and identified tryptic peptides from diel cycle and iron limitation of Crocosphaera watsonii: PNAS2011Supplemental (also available on PNAS site as an open access document).
B12 Limitation of Marine Diatoms
Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar T, Bai J, Saito MA. 2012. Impact of Cobalamin Starvation on Diatom Molecular Physiology and the Identification of a Novel Cobalamin Acquisition Protein. Proc. Natl. Acad. Sci.
Spectral Counting results and identified peptides under B12 and iron colimitation: Supplemental Proteomics Dataset
Phosphorus limitation in Thalassiosira pseudonana
ST Dyhrman, BD Jenkins, TA Rynearson, MA Saito, ML Mercier, H Alexander, LP. Whitney, A Drzewianowski, VV Bulygin, EM Bertrand, Z Wu, C Benitez-Nelson, A Heithoff. 2012 Coordination in the transcriptome and proteome of the diatom Thalassiosira pseudonana reveals a diverse phosphorus stress response. PLoS One.
Supplemental datasets
Peptides from Thalassiosira pseudonana under phosphorus-limited and replete conditions: Dyhrman_PLOSONE2012_Table4Peptides
Spectral count and corresponding transcriptome data from phosphorus-limited and replete Thalassiosira pseudonana: Dyhrman_PLOSONE_Table5SupplementalSpectralCounts
Phosphorus limitation in Aureococcus anophagefferens
Louie L. Wurch, Erin M. Bertrand, Mak A. Saito, Benjamin A.S. Van Mooy and Sonya T. Dyhrman. 2011. Proteome changes driven by phosphorus stress and recovery in the brown tide-forming alga, Aureococcus anophagefferens. PLoS One.
Supplemental Proteomics Datasets
Identified peptides: Wurch_TableS1Peptides
Protein spectral count data and corresponding transcriptome results: Wurch_TableS2SpectralCounts
Metaproteomic Datasets
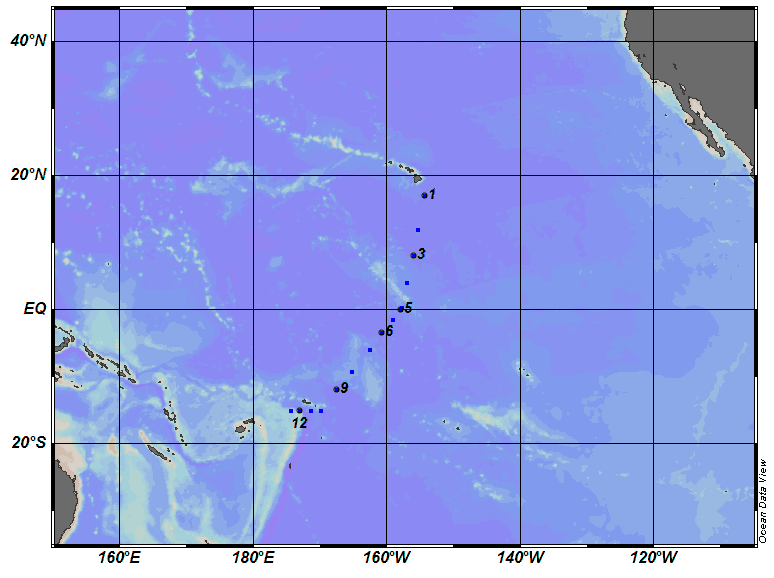
METZYME Expedition Hawaii-Samoa Nov-Dec 2011 R/V Kilo Moana
Samples analyzed by LC-MS (Thermo LTQ) for initial proteomic discovery work followed by analysis in X!!Tandem. Currently identified peptides dataset on station links below (Version July 10, 2013). These samples contain filtered material between 0.2 microns to 3.0 microns in size. Further high resolution and quantitative LC-MS analyses are underway. Funded by the Gordon and Betty Moore Foundation and Chemical Oceanography National Science Foundation.
|
METZYME
|
KM1128
|
|||
|
KM1128
|
OCTOBER -NOVEMBER 2011
|
|||
|
Station
|
Station Start Date
|
Long E
|
Lat N
|
Depth (M)
|
|
10/3/2011
|
205.60
|
17.00
|
50
|
|
|
10/3/2011
|
205.60
|
17.00
|
90
|
|
|
10/3/2011
|
205.60
|
17.00
|
120
|
|
|
10/3/2011
|
205.60
|
17.00
|
200
|
|
|
10/3/2011
|
205.60
|
17.00
|
300
|
|
|
10/3/2011
|
205.60
|
17.00
|
400
|
|
|
10/3/2011
|
205.60
|
17.00
|
600
|
|
|
10/8/2011
|
204.00
|
8.00
|
40
|
|
|
10/8/2011
|
204.00
|
8.00
|
60
|
|
|
10/8/2011
|
204.00
|
8.00
|
120
|
|
|
10/8/2011
|
204.00
|
8.00
|
150
|
|
|
10/8/2011
|
204.00
|
8.00
|
200
|
|
|
10/8/2011
|
204.00
|
8.00
|
250
|
|
|
10/8/2011
|
204.00
|
8.00
|
300
|
|
|
10/8/2011
|
204.00
|
8.00
|
500
|
|
|
10/8/2011
|
204.00
|
8.00
|
550
|
|
|
10/8/2011
|
204.00
|
8.00
|
600
|
|
|
10/8/2011
|
204.00
|
8.00
|
800
|
|
|
10/13/2011
|
202.01
|
0.00
|
20
|
|
|
10/13/2011
|
202.01
|
0.00
|
50
|
|
|
10/13/2011
|
202.01
|
0.00
|
80
|
|
|
10/13/2011
|
202.01
|
0.00
|
120
|
|
|
10/13/2011
|
202.01
|
0.00
|
200
|
|
|
10/13/2011
|
202.01
|
0.00
|
300
|
|
|
10/13/2011
|
202.01
|
0.00
|
400
|
|
|
10/13/2011
|
202.01
|
0.00
|
500
|
|
|
10/13/2011
|
202.01
|
0.00
|
600
|
|
|
10/17/2011
|
199.23
|
-3.50
|
40
|
|
|
10/17/2011
|
199.23
|
-3.50
|
80
|
|
|
10/17/2011
|
199.23
|
-3.50
|
200
|
|
|
10/19/2011
|
194.64
|
-9.25
|
40
|
|
|
10/19/2011
|
194.64
|
-9.25
|
70
|
|
|
10/19/2011
|
194.64
|
-9.25
|
200
|
|
|
10/20/2011
|
192.44
|
-12.00
|
40
|
|
|
10/20/2011
|
192.44
|
-12.00
|
70
|
|
|
10/20/2011
|
192.44
|
-12.00
|
380
|
|
|
10/23/2011
|
186.90
|
-15.00
|
40
|
|
|
10/23/2011
|
186.90
|
-15.00
|
120
|
|
|
10/23/2011
|
186.90
|
-15.00
|
300
|
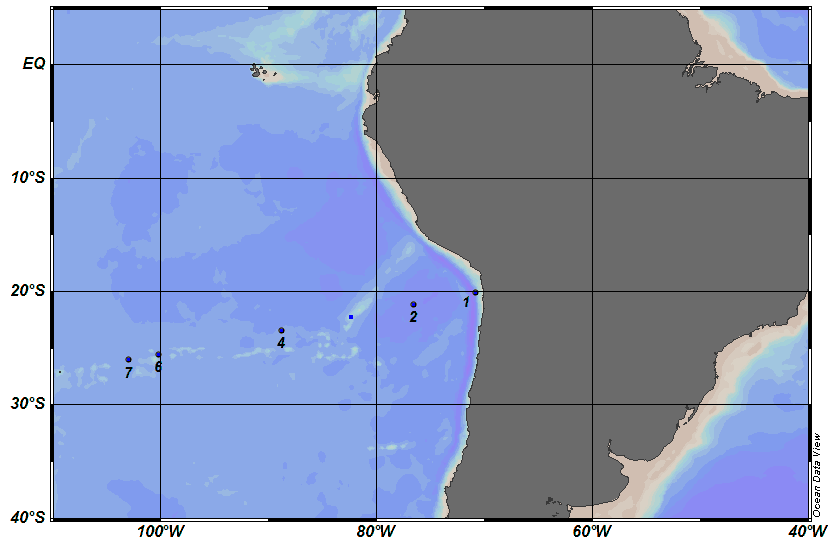
BIG RAPA Chile-Easter Island Expedition R/V Melville 2010
Samples analyzed by LC-MS (Thermo LTQ) for initial proteomic discovery work followed by analysis in X!!Tandem. Currently identified peptides dataset (in preparation). These samples contain filtered material between 0.2 microns to 3.0 microns in size. Further high-resolution and quantitative LC-MS analyses are underway. Funded by the Gordon and Betty Moore Foundation and Center for Microbial Oceanography Research and Eduction (C-MORE, National Science Foundation).
| BIG RAPA | MV1015 | |||
| Nov-Dec 2010 | ||||
| Station | Station Start Date | Long E | Lat N | Depth (M) |
| 1 | 11/22/2010 | 289.20 | -20.08 | 25 |
| 2 | 11/25/2010 | 283.43 | -21.18 | 48 |
| 2 | 11/25/2010 | 283.43 | -21.18 | 110 |
| 2 | 11/25/2010 | 283.43 | -21.18 | 800 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 20 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 100 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 200 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 375 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 500 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 650 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 800 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 1000 |
| 4 | 11/29/2010 | 271.24 | -23.46 | 1500 |
| 6 | 12/6/2010 | 259.86 | -25.55 | 10 |
| 6 | 12/6/2010 | 259.86 | -25.55 | 20 |
| 6 | 12/6/2010 | 259.86 | -25.55 | 60 |
| 6 | 12/6/2010 | 259.86 | -25.55 | 120 |
| 6 | 12/6/2010 | 259.86 | -25.55 | 250 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 20 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 75 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 175 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 250 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 400 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 500 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 800 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 1100 |
| 7 | 12/7/2010 | 257.10 | -26.05 | 1900 |
Related Files
» Metzyme_S1
» Metzyme_S3
» Metzyme_S5
» Metzyme_S6
» Metzyme_S8
» Metzyme_S9
» Metzyme_S12
Computational Proteomics Tools
Redundant Tryptic Peptide Scripts
These scripts provide tools and documentation for detecting redundant tryptic peptides between multiple genomes. These tools are intended to assist proteomics scientists understand the potential for identifying unique and shared tryptic peptides in metaproteomic data. Github repository link.