Innovation
The Woods Hole COHH is innovative in the research proposed, the technologies employed, our current findings that have led to the research, the broad and perhaps global implications for HAB biology, the models to predict HAB events in a changing ocean, and the new understanding and implications of developmental neurotoxic mechanisms we are uncovering. A unique and notable aspect of the Program is that these studies employ a variety of in situ sensor technologies, including new instrumentation and deployment approaches developed by the applicants, to detect and characterize fully the key mechanisms regulating Alexandrium and Pseudo- nitzschia population dynamics in nature, and their response to climatic variation.
Similarly, the studies of novel mechanisms of developmental effects of toxins, which we have uncovered, employ modern technologies, including transgenic zebrafish lines, high-speed video analysis of behavior, and epitranscriptomics.

Environmental Sample Processor (ESP)
The Environmental Sample Processor (ESP) is a novel robotic system that remotely collects samples of seawater, filters out cells, breaks them apart, and then analyzes the DNA and indentifying chemicals resulting from the cellular decomposition. The data is then transmitted back to scientists in the lab. Moored at sea, the ESP allows for the real time monitoring of ocean conditions, allowing scientists to detect harmful algal blooms (HAB) as they begin. This provides an effective early warning system and can also collect in situ data needed to improve forecast models. Furthermore, the ESP is a versatile system and can be calibrated to detect not only algae but bacterial or larval organisms as well. Ongoing research is also developing capabilities for HAB toxin measurements on the ESP. This makes the ESP an ideal system for a variety of different scientific applications. Developed by the Monterey Bay Aquarium Research Institute and built by McLane Research Laboratories, further information regarding the ESP can be found below.
Related Links
- Scientists Use "ESP" to Track Harmful Algae
Oceanus Magazine - Environmental Sample Processor
McLane Research Laboratories - Environmental Sample Processor
Monterey Bay Aquarium Research Institute - Anderson Lab
Woods Hole Oceanographic Instituion - Doucette Lab
National Oceanic and Atmospheric Administration


Imaging FlowCytobot (IFCB)
The Imaging FlowCytobot (IFCB) is a system that utilizes video and flow cytometric technology to photograph and classify a variety of microorganisms, revealing to scientists on shore the growth and decline of diverse plankton populations. The system acts as a type of submerged microscope, analyzing microorganisms on a continuous basis, while simultaneously transmitting this information to the shore station in real time. Capable of long-term deployment, the FlowCytobot offers a comprehensive and efficient monitoring of microorganism populations. Through automated image classification methods, phytoplankton and other microorganisms are identified and counted providing unprecidented detail on complexities of plankton dynamics.
Related Links
- Cytobot Gives Early Red Tide Warning
Oceanus Magazine - Profile of FlowCytobot
Woods Hole Oceanographic Institution - Sosik Laboratory
Woods Hole Oceanographic Institution - Imaging FlowCytobot
McLane Research Laboratories
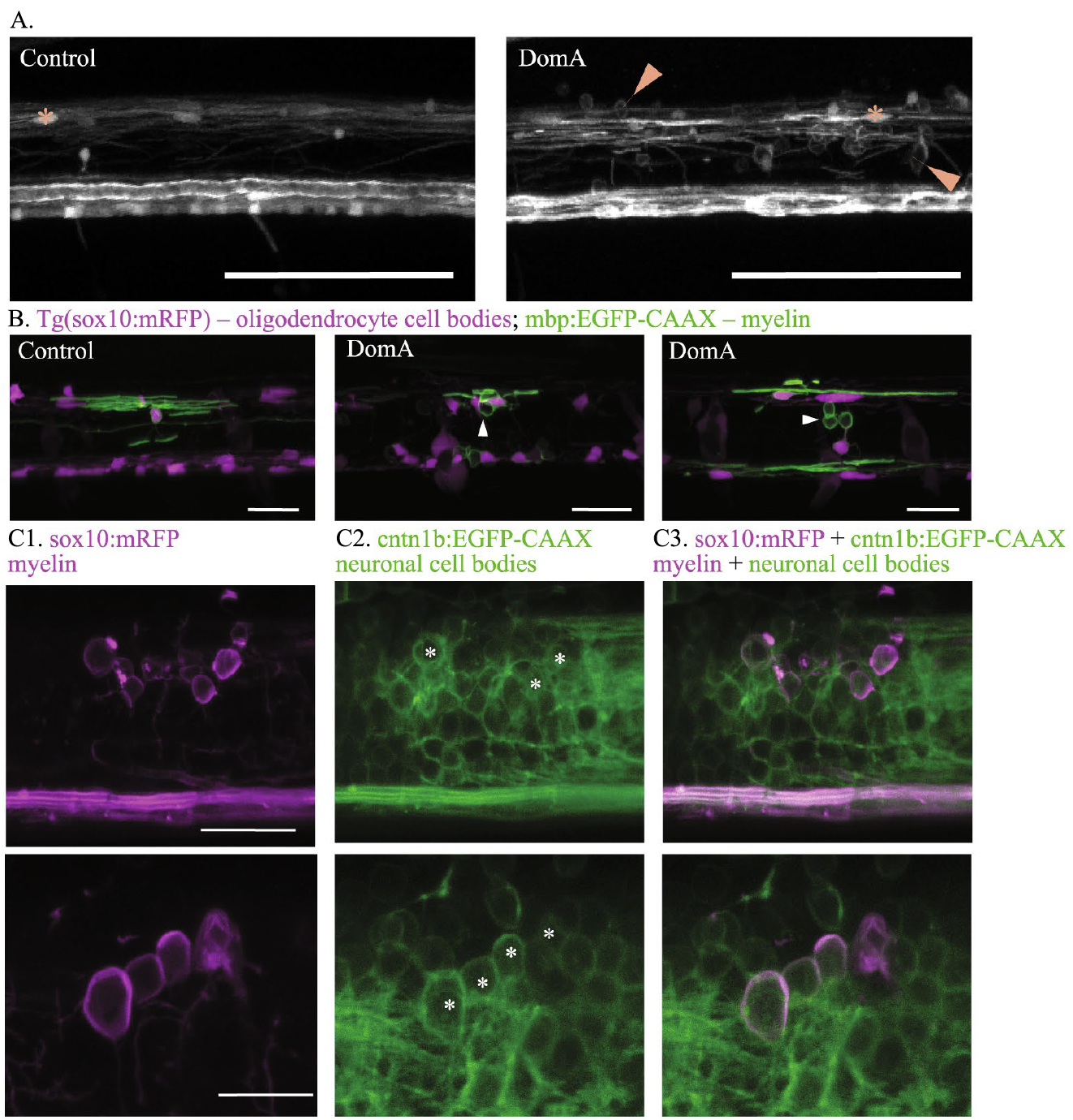
Transgenic zebrafish
One of the advantages of the zebrafish embryo model is the ability to generate transgenic lines in which specific cell types are labeled with fluorescent markers such as green fluorescent protein (GFP; which was first cloned by WHOI researcher Doug Prasher). We are using a variety of transgenic lines in which neurons and glial cells (including myelinating oligodendrocytes) are labeled with fluorescent tags. With these lines, we use image analysis and time-lapse imaging in vivo to determine the effects of exposure to toxins such as domoic acid or saxitoxin, or toxicants such as PCBs, on the developmental trajectory and function of these cells. For examples, please see the papers authored by Panlilio et al (2020, 2021, 2023) in the publications list.
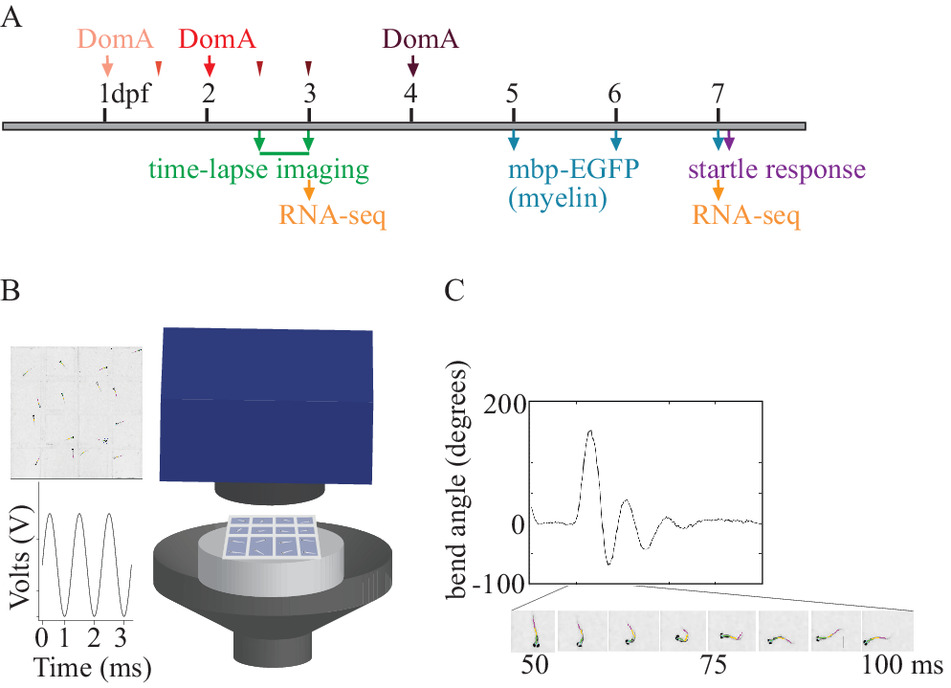
High-speed video analysis of startle behavior
Startle responses in teleosts are elicited by sudden stimuli. Auditory/vibrational stimuli can lead to one of two types of startle responses — a short-latency c-type (SLC) startle response (<14 ms) or a long-latency c-type (LLC) startle response (>14 ms). These two types of startle response have different latencies, distinct kinematics, and are driven by separate underlying neural circuits. The startle response circuits are well known in zebrafish, making it possible to link startle behavioral data to underlying structural and cellular targets (see Transgenic zebrafish, above). High-speed videography is required to differentiate these two types of startle responses. See Panlilio et al (2020, 2021) and Brun et al (2021) for details of the assay and its use to examine mechanisms of developmental neurotoxicity.