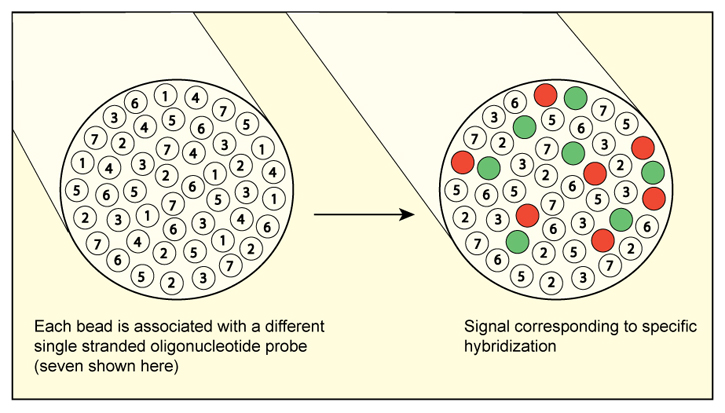
Fiber Optic Microarray to Detect and Enumerate Harmful Algal Bloom Species
Harmful algal blooms (HABs) have become a serious threat to coastal resources worldwide, causing impacts ranging from the contamination of seafood products with potent toxins to widespread mortalities of wild and farmed fish and other marine animals. In the U.S., thousands of miles of coastline are affected, including many estuaries and sounds within the Gulf of Maine (GOM). The most serious HAB problem in the GOM is paralytic shellfish poisoning, a syndrome caused by human ingestion of shellfish that accumulate toxins from the dinoflagellate Alexandrium fundyense. Several other HAB species of concern are commonly found within the GOM, in particular diatoms in the genus Pseudo-nitzschia, which cause amnesic shellfish poisoning, and the dinoflagellate Alexandrium ostenfeldii, which produces a newly discovered toxin family called spirolides. As the threat from HABs has expanded worldwide, new approaches to monitoring have become necessary - in particular monitoring the plankton for HAB species. Coastal managers will need rapid yet sensitive methods that can accurately identify and enumerate the multiple HAB species that may be present in a single water sample. The coastal and estuarine waters of the Wells National Estuarine Research Reserve are home to all three of the HAB species, making it the ideal location for the field aspects of this project.
Our proposed project employs an innovative approach to HAB cell enumeration - fiber optic genosensors. The project team has successfully developed an operational, multi-species fiber optic array, and now the overall project goal is to refine and validate a rapid and accurate optical fiber-based technology for HAB cell detection and enumeration that can be used in both laboratory and field settings. Specific objectives are to:
- Redesign A. ostenfeldii probe(s) to lower detection limits;
- Design and test probes for toxic Pseudo-nitzschia species from the Gulf of Maine;
- Design and test a second probe pair for each species, to incorporate redundancy into the array;
- Refine hybridization conditions to reduce processing time;
- Fully test the 3-species multiplex array using mixed cultures, and simulated and natural field samples;
- Work with managers responsible for coastal monitoring to determine desired detection limits, precision, and other operational characteristics for the assay and expected instrumentation.
This project addresses a significant need in the field of red tide and HAB monitoring and coastal management: methods that accelerate and simplify enumeration of HAB species. This technology can readily be adapted to target other HAB species as well as microbial pathogens and microorganisms of many types. Furthermore, it is highly amenable to automation, bringing us closer to the goal of an early warning system of moored instruments capable of detecting organisms that threaten public health and fisheries operations.
Relevant Publications
Ahn, S., D.M. Kulis, D.L. Erdner, D.M. Anderson and D.R. Walt. 2006. Fiber-optic microarray for simultaneous detection of multiple harmful algal bloom species. Applied and Environmental Microbiology 72(9):5742-5749.
Anderson, D.M., D. Kulis, D. Erdner, S. Ahn, and D. Walt. 2006. Fibre optic microarrays for the detection and enumeration of harmful algal bloom species. African Journal of Marine Science 28(2): 231-25.
Additional Information
CICEET: The Cooperative Institute for Coastal and Estuarine Environmental Technology
» Visit website
Two if by Sea: Scientists Join Forces in Search for Faster Detection of Harmful Algal Blooms
» Visit website
David Walt: Department of Chemistry, Tufts University
» Visit website