Coupling microbial ecology to the biogeochemical cycling of dissolved organic matter in the oligotrophic ocean.
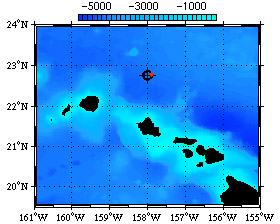
Dissolved organic matter (DOM) is one of the largest and most dynamic carbon reservoirs on the planet, and serves as the center at the marine microbial food web. Spectral analyses show that a good portion of DOM is comprised of a closely related family of polysaccharides with a remarkably uniform composition that is conserved across all major ocean basins. DOM polysaccharides collected at Station ALOHA (Fig. 1) are chemically and spectrally indistinguishable from DOM collected in the North Atlantic Ocean (off Bermuda). The presence of such a ubiquitous, specific, and abundant component of DOM provides us with a chemically well defined and biogeochemically important substrate that we can target for microbial-DOM studies.
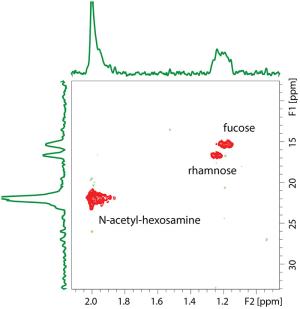
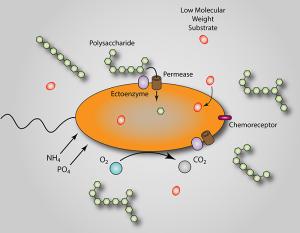
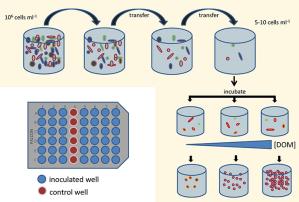
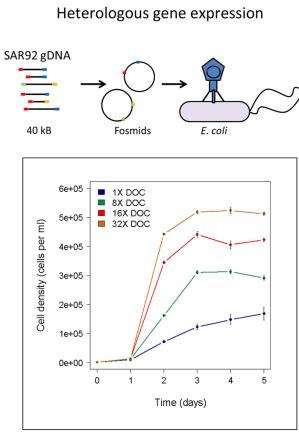
The metabolic and biogeochemical pathways responsible for DOM cycling are “black boxes” in the context of the biology and chemistry of marine carbon cycling. In collaboration with Ed DeLong’s group at MITand with funding from the Gordon and Betty Moore Foundation we are working to isolate, cultivate and characterize microbes that drive DOM polysaccharide cycling, and to develop model systems to explore the physiology, ecology and biogeochemistry of DOM turnover in nutrient-poor waters. DOM polysaccharides are isolated (Figs. 2&3), purified, characterized (Fig. 4), and used as substrates in high throughput culture screens (Fig. 5), which are profiled using genomic and transcriptomic approaches (Figs. 6, 7). Our approach leverages the microbiology to elucidate details of DOM-degradation biochemistry, and conversely uses DOM chemical characterization and manipulation to test hypotheses microbial diversity and metabolism in open ocean oligotrophic gyres.
Elemental, isotopic, spectral, and chromatographic data along with analytical protocols can be found within the subpage link at upper left