Metabolic profiling and bioactive organic compounds in laboratory pure cultures and seawater
Microbial ecosystems across large expanses of the global ocean are characterized by specific classes of marine phytoplankton. For example, diatoms are often abundant in high nutrient coastal waters, while cyanobacteria often dominate the nutrient-poor oligotrophic gyres. Bacteria live in association with these communities, consuming the dissolved organic carbon, nitrogen, and phosphorus that are by-products of marine primary production. In return, bacteria supply phytoplankton with essential organic nutrients such as vitamins and organically bound trace metals. Numerous studies have shown a loose symbiosis between microbial photoauto- and heterotrophs that is mediated in part by an exchange of dissolved organic matter. But of the tens to hundreds of thousands of organic compounds dissolved in each drop of seawater, what are the compounds that matter most to marine microbes?
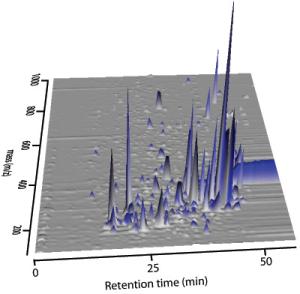
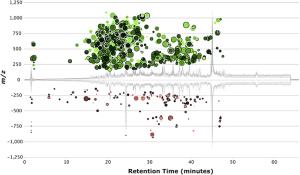
To answer this question, we are characterizing dissolved organic matter in a number of model marine photoautotrophs with sequenced genomes. With support from the Gordon and Betty Moore Foundation, and from the NSF-CMORE Science and Technology Center, we grow bacteria-free, pure cultures of marine algae and cyanobacteria (Fig. 1), recover the DOM (Figs. 2, 3), and perform detailed chemical analyses by combined high pressure liquid chromatography-mass spectrometry (HPLC-MS; Fig. 4). HPLC separates the complex mix of DOM released by model microbes by their relative polarity (retention time), while MS separates constituents by their mass. The combination of retention time and mass define a “feature” which is either a pure organic compound or simple mixture of closely related compounds (Fig. 5). We can compare features between cultures to discover unique or common compounds that can be targeted for further chemical analyses or used experimentally to screen for biological activity (Fig. 6). In a recent experiment we screened DOM from Prochlorococcus against a suite of bacteria that were isolated from Hawaii. We find only specific bacteria grow in response to Prochlorococcus DOM, and we are now searching through our database of Prochlorococcus isolates to identify specifically which compounds stimulate uptake. In parallel, we are analyzing the bacterial transcriptome to better understand what pathways the bacteria use to metabolize DOM.