Dispatch 22: It’s a Small World, After All
Sara Palestini
October 6, 2023
Dispatch 22: It’s a Small World, After All
Greetings All! My name is Sara Palestini, and I am a first-year master’s student at Concordia University. I am part of the science personnel aboard the CCGS Louis S. St-Laurent, studying the microbial diversity of the Beaufort Gyre.
Fungi, bacteria, archaea, and protists are all members of the microscopic organisms that dominate the planet. Microbial life also represents the greatest biological diversity on Earth. Despite a fascinating variety of metabolisms, and having colonized every environment known to us, they are still far less understood than larger forms of life, like animals and plants. Many microbes are resistant to culturing and so those that have been most thoroughly studied are those that are easily cultured in a lab setting -with greatest effort placed into the study of human pathogens. Microbial taxa are also alarmingly abundant, and thousands of species might exist in a single drop of water. The challenge presented to microbial ecologists then is how to study a microbial community when traditional methods won’t do. This can be accomplished through genomics. Studying the genome of a microbe provides insights into their metabolism, adaptations to their environment, and their evolution. Genetic sequencing and genome assembly techniques have become more effective, inexpensive, and capable of reconstructing a large number of genomes from an environmental sample with reasonable confidence; they are therefore great and accessible tools to explore the microbial world. My job onboard is to collect seawater samples from across the water column and filter them, thus capturing the microbial community at a given depth and time. Back home, a catalogue of the community’s genomic data (metagenome) can be assembled, and individual genomes can be reconstructed into what we call MAGs (metagenome assembled genomes).
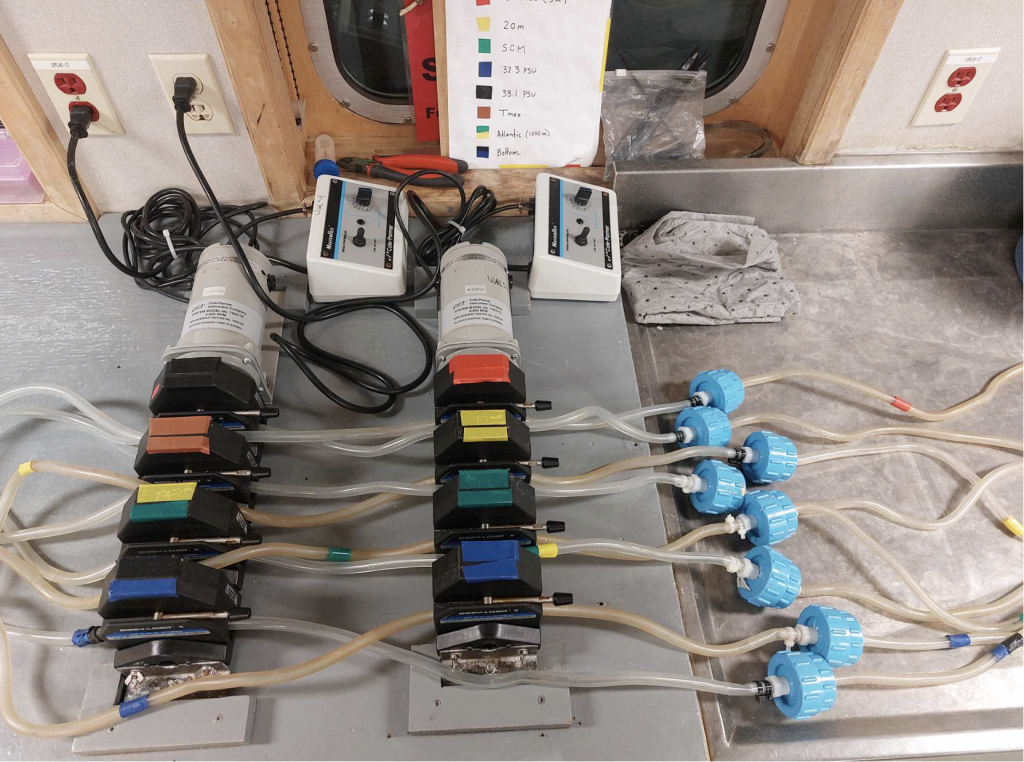
From the bottom water to the surface, each layer in the water column is a unique habitat which hosts a different cast of characters with varying metabolic activity and niches. Water from the rosette is collected into individual carboys (Figure 5) and filtered through identical filtration systems. Each filtration system is comprised of two filters; the first is a 47mm diameter membrane (Figures 3-4) with a pore size of 3 micrometers (0.003mm), which is placed gently into a cleaned filter holder and sealed, and a second filter with a smaller 0.2-micrometer (0.0002mm) pore size inside of a sterile tube (Figure 2) connected to the end of the tubing by a luer-lock. As water runs from the carboys it is first intercepted by the 3-micrometer filter, which retains mostly multi-celled organisms and larger eukaryotic cells and allows many prokaryotes -like bacteria and archaea- to pass while the second filter retains them. These filters are connected by tubing to the carboys and a peristaltic pump (Figure 1) is used to push water along. The carboys used are either 5L or 20L but can hold 7L and 28L respectively when filled to the top. The larger carboys are used at only a select few stations. The samples collected from these carboys not only go towards generating MAGs but also towards recovering RNA and analyzing the expression of certain genes. When used in conjunction with the metagenomic data we collected we can infer what activities a population within the sample may be carrying out. Once the carboys are done emptying, the filters are contained and preserved using RNAlater™ solution, which doubly deactivates the enzymes which breakdown genetic material and stabilizes genetic material for preservation. From there the sealed vials containing the filters are placed in freezer safe boxes and sent off to the freezer downstairs, where they will await the trip home.